Abstract
Extranodal natural killer (NK)/T cell lymphoma, nasal type (ENKTL) is an aggressive, rare form of Epstein-Barr virus (EBV)-positive non-Hodgkin lymphoma that typically presents in the naso/oropharynx and is associated with high rates of systemic relapse and poor survival. The neoplastic cells in ENKTL are usually of NK cell origin, yet how they relate to the distinct stages that define normal NK cell development has not been fully described. We have established a model of human NK cell development where early NK cell precursors progress through sequential stages of maturation within mucosal associated lymphoid tissues (MALT) and are terminally differentiated in the peripheral blood (PB) (Freud et al. Cell Reports, 2016). Here we aimed to characterize neoplastic NK cells from ENKTL patients within this NK cell developmental framework.
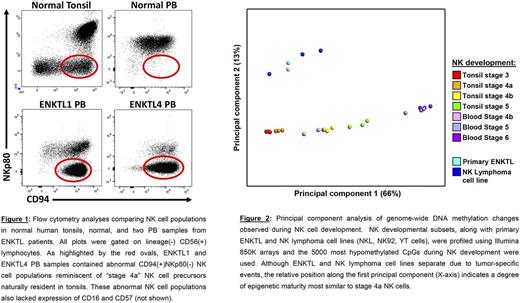
In this study we performed flow cytometry analyses, genome-wide DNA methylation profiling, and RNA sequencing of normal NK cell developmental subsets from 12 healthy donor PB and 12 tonsil specimens and of neoplastic NK cells from PB, bone marrow (BM), or cerebrospinal fluid of 11 patients with relapsed ENKTL. While all 11 of the samples contained NK cells, four of the 11 ENKTL patient samples contained distinctly abnormal NK cell populations that expressed CD56 and CD94 but lacked CD16, CD57, and NKp80. We noted that the recurring CD16(-)CD56(+)CD57(-)CD94(+)NKp80(-) phenotype of these cells was similar to that of MALT-resident "stage 4a" NK cell precursors that are rarely detected in the PB (Figure 1).
To further investigate the ENKTL phenotype, we sorted two of the abnormal patient-derived CD16(-)CD56(+)CD57(-)CD94(+)NKp80(-) NK cell populations to >98% purity. Real-time quantitative PCR for the EBNA1 gene confirmed that the sorted cells were EBV(+). We then compared the ENKTL cells to normal NK cell developmental intermediates from healthy donors using genome-wide DNA methylation profiling and global gene expression analysis by RNA sequencing. DNA methylation analyses of human tonsil- and PB-derived NK cell developmental subsets revealed a broad, stage-wise acquisition of an NK cell-specific epigenetic program involving targeted gain and loss of methylation enriched in promoters and enhancer regions of genes with known relevance for NK cell development and function. Hypomethylation during development was found to be highly associated with T-BET, EOMES, ETS, KLF and RUNX family transcription factor binding sites, whereas hypermethylation was associated with RORγt, NFkB, and TCF/E2A ( P <1x10-10). Expression of specific transcription factors and NK cell-related genes mirrored observed methylation changes. These analyses also revealed that the DNA methylation patterns of the purified ENKTL cells were most similar to those of MALT-derived stage 4a NK cells (Figure 2).
Lastly, we transplanted purified ENKTL cells into immune deficient NOD- scid IL2Rgammanull mice. Whereas normal donor tonsil-derived stage 4a precursor cells underwent terminal differentiation in vivo, the transplanted ENKTL cells maintained the stage 4a-like phenotype and did not show evidence of terminal NK cell differentiation despite extensive expansion and infiltration of numerous organs. Global DNA methylation patterns also remained stable between the primary ENKTL cells and cells collected from the BMs and spleens of engrafted mice. Collectively, the flow cytometry, RNA expression, and epigenetic data show that the CD16(-)CD56(+)CD57(-)CD94(+)NKp80(-) ENKTL phenotype in the patient samples examined is stable and most similar to that of MALT-resident stage 4a NK cell precursors. These findings provide new insight into the biological nature of ENKTL that may guide development of novel therapeutic approaches for this aggressive disease.
Brammer: Celgene: Research Funding. Baiocchi: Prelude therapeutics: Research Funding; essanex: Research Funding; Theravectys: Consultancy; viracta: Membership on an entity's Board of Directors or advisory committees, Research Funding. Porcu: Kura: Research Funding; Miragen: Research Funding; Kiowa: Research Funding; Galderma: Research Funding; Innate Pharma: Research Funding; Tetralogic: Research Funding; Celgene: Research Funding; Cell Medica: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal